Recently, I did my machine learning assignment, and I want to build a neural network with keras on jupyter notebook. As a result, even the simplest layer of neural network couldn’t work. What’s more strange is that I ran the iris data set first, and there was no problem, but I used the fashion MNIST given by my teacher to prompt the server to hang up and restart as soon as I ran it. What’s more strange is that there’s no problem with the same code running on the students’ computers, which makes me think that my MacBook is too old and the configuration is too low, so I almost want to buy a new computer & gt_& lt;
After several times of debugging by ml teacher in class today, the problem has been solved perfectly, worthy of CMU God( Call Prof strongly here, though he doesn’t understand Chinese & gt& lt;) Because I haven’t learned Python for a long time and I’m not familiar with it. After this time, I’ve learned many new skills ✌️
The complete code of the problem is as follows, which is to use keras to realize logistic region. It is a simple one layer network, but every time it runs to the last line, the server will hang up, and then restart kernel
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA, FastICA
from sklearn.linear_model import LogisticRegression
from keras.models import Sequential
from keras.layers import Dense, Activation, Conv2D
from keras.utils import to_categorical
from keras.datasets import fashion_mnist
(x3_train, y_train), (x3_test, y_test) = fashion_mnist.load_data()
n_classes = np.max(y_train) + 1
# Vectorize image arrays, since most methods expect this format
x_train = x3_train.reshape(x3_train.shape[0], np.prod(x3_train.shape[1:]))
x_test = x3_test.reshape(x3_test.shape[0], np.prod(x3_test.shape[1:]))
# Binary vector representation of targets (for one-hot or multinomial output networks)
y3_train = to_categorical(y_train)
y3_test = to_categorical(y_test)
from sklearn import preprocessing
scaler = preprocessing.StandardScaler()
x_train_scaled = scaler.fit_transform(x_train)
x_test_scaled = scaler.fit_transform(x_test)
n_output = y3_train.shape[1]
n_input = x_train_scaled.shape[1]
nn_lr = Sequential()
nn_lr.add(Dense(units=n_output, input_dim= n_input, activation = 'softmax'))
nn_lr.compile(optimizer = 'sgd', loss = 'categorical_crossentropy', metrics = ['accuracy'])
Since Jupyter Notebook just keeps restarting the kernel and there is no error message, so people can’t get started. But after the teacher prompted the original Jupyter Notebook automatically opened when the terminal will record information on the run (the first time a white person found.) The following is the detailed process of kerter abort and restart and the reasons for it.
[I 22:11:54.603 NotebookApp] Kernel interrupted: 7e7f6646-97b0-4ec7-951c-1dce783f60c4
[I 22:13:49.160 NotebookApp] Saving file at /Documents/[Rutgers]Study/2019Spring/MACHINE LEARNING W APPLCTN LARGE DATASET/hw/Untitled1.ipynb
2019-03-28 22:13:49.829246: I tensorflow/core/platform/cpu_feature_guard.cc:141] Your CPU supports instructions that this TensorFlow binary was not compiled to use: SSE4.1 SSE4.2 AVX AVX2 FMA
2019-03-28 22:13:49.829534: I tensorflow/core/common_runtime/process_util.cc:69] Creating new thread pool with default inter op setting: 4. Tune using inter_op_parallelism_threads for best performance.
OMP: Error #15: Initializing libiomp5.dylib, but found libiomp5.dylib already initialized.
OMP: Hint: This means that multiple copies of the OpenMP runtime have been linked into the program. That is dangerous, since it can degrade performance or cause incorrect results. The best thing to do is to ensure that only a single OpenMP runtime is linked into the process, e.g. by avoiding static linking of the OpenMP runtime in any library. As an unsafe, unsupported, undocumented workaround you can set the environment variable KMP_DUPLICATE_LIB_OK=TRUE to allow the program to continue to execute, but that may cause crashes or silently produce incorrect results. For more information, please see http://www.intel.com/software/products/support/.
[I 22:13:51.049 NotebookApp] KernelRestarter: restarting kernel (1/5), keep random ports
kernel c1114f5a-3829-432f-a26a-c2db6c330352 restarted
There is another way to get a similar message by copying the code into ipython, so the final error located is
OMP: Error #15: Initializing libiomp5.dylib, but found libiomp5.dylib already initialized.
Google it, there is a very detailed discussion post on github, but the owner was running XGBoost when he encountered this problem, which reminds me that the winter break installation of XGBoost did go through a very tortuous process, and may have accidentally downloaded a file to a different path repeatedly, so the program loaded the package when there was a conflict. The post provides several possible causes and solutions.
1. uninstall clang-omp
brew uninstall libiomp clang-omp
as long as u got gcc v5 from brew it come with openmp
follow steps in:
https://github.com/dmlc/xgboost/tree/master/python-package
Tried uninstalling xgboost and installing it again, then uninstalling clang-omp, got an error
No such keg: /usr/local/Cellar/libiomp
pip uninstall xbgoost
pip install xgboost
brew uninstall libiomp clang-omp
2. Run directly in jupyter notebook
# DANGER! DANGER!
import os
os.environ['KMP_DUPLICATE_LIB_OK']='True'
The teacher said that this command allows the system to ignore the problem of package conflict and choose a package to use. Try this method is really effective, but it is very dangerous, extremely not recommended
3. Find the duplicate libiomp5.dylib file and delete one of them
Two files are found in finder, which are in ~/ anaconda3 /lib and ~/anaconda3 / lib / Python 3.6 / site packages /_ solib_ darwin/_ U@mkl_ Udarwin_ S_ S_ Cmkl_ Ulibs_ Udarwin___ Uexternal_ Smkl_ Udarwin_ Slib (????) But I’m not sure which one should be deleted. I think it’s dangerous. I can’t run if I delete the wrong one
4. OpenMP conflict
Hint: This means that multiple copies of the OpenMP runtime have been linked into the program
According to hint in the prompt message, I searched tensorflow OpenMP. OpenMP is a multi thread parallel programming platform, tensorflow seems to have its own parallel computing architecture, and can not use OpenMP (see) https://github.com/tensorflow/tensorflow/issues/12434 )
5. Unload nomkl
I had the same error on my Mac with a python program using numpy, keras, and matplotlib. I solved it with ‘conda install nomkl’.
This is the last effective way! The full name of nomkl is math kernel library (MKL) optimization. It is a module developed by Interl to speed up mathematical operations. MKL can be used automatically by installing package through CONDA. For more details, please refer to the official document of anaconda
To opt out, runcondainstallnomkland then usecondainstallto install packages that would normally include MKL or depend on packages that include MKL, such asscipy,numpy, andpandas.
Maybe there are some conflicts in the update of package such as numpy. After installing nomkl, it was solved miraculously. Later, it tried to unload MKL, and the program is still running normally.. The unload command is as follows:
conda remove mkl mkl-service
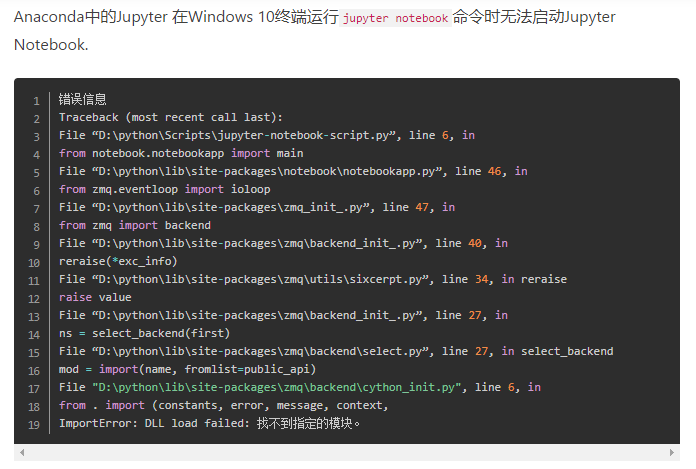
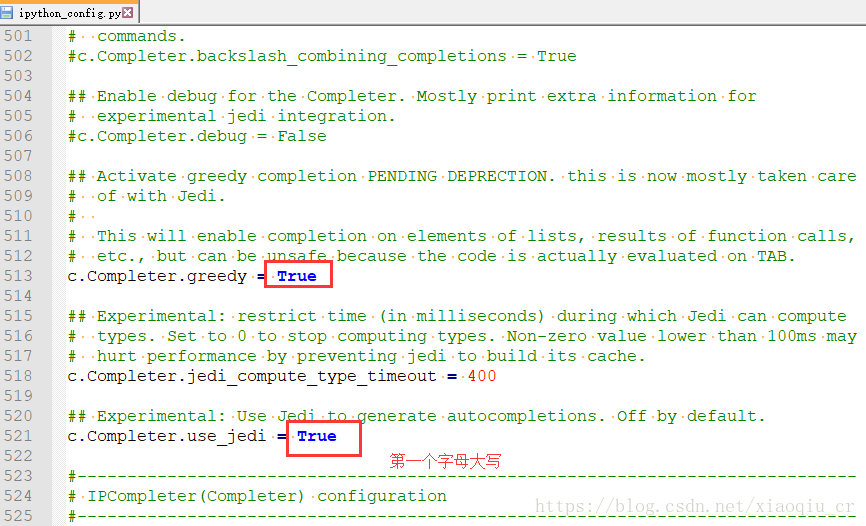
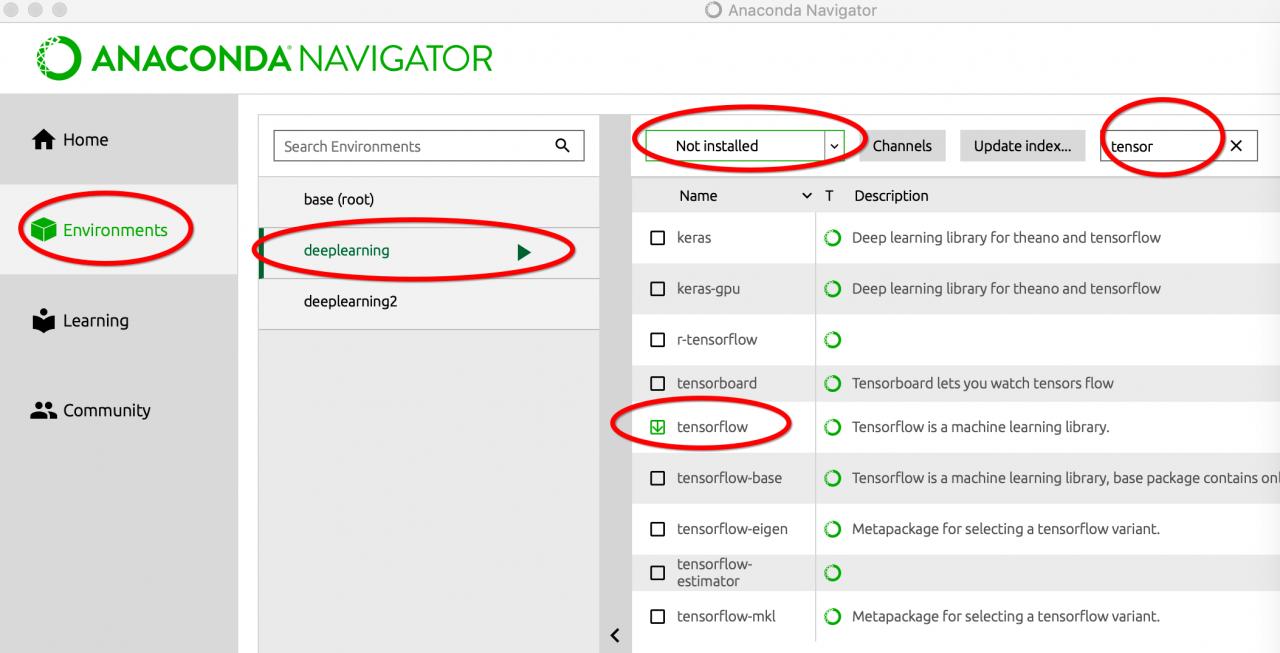
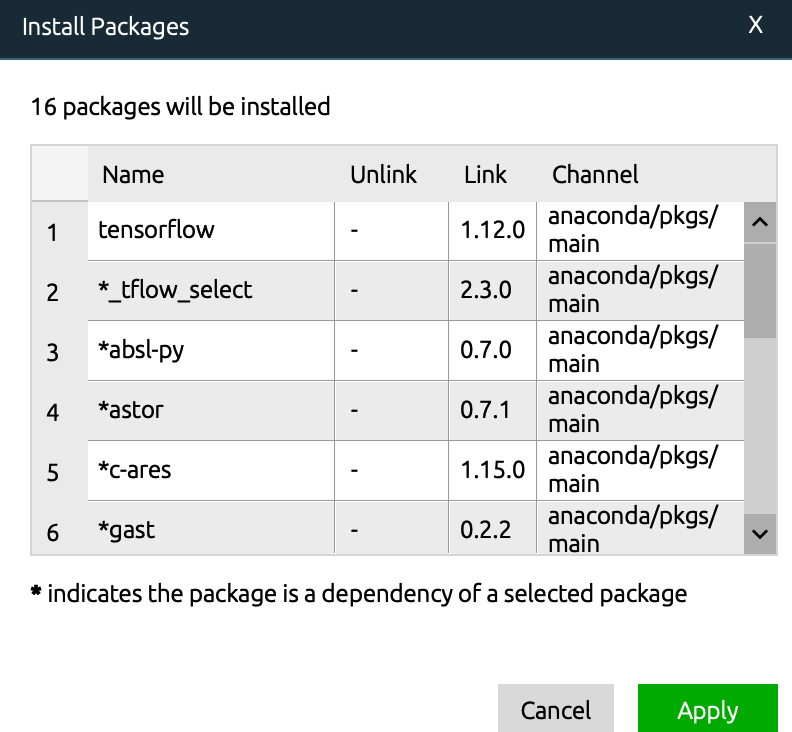
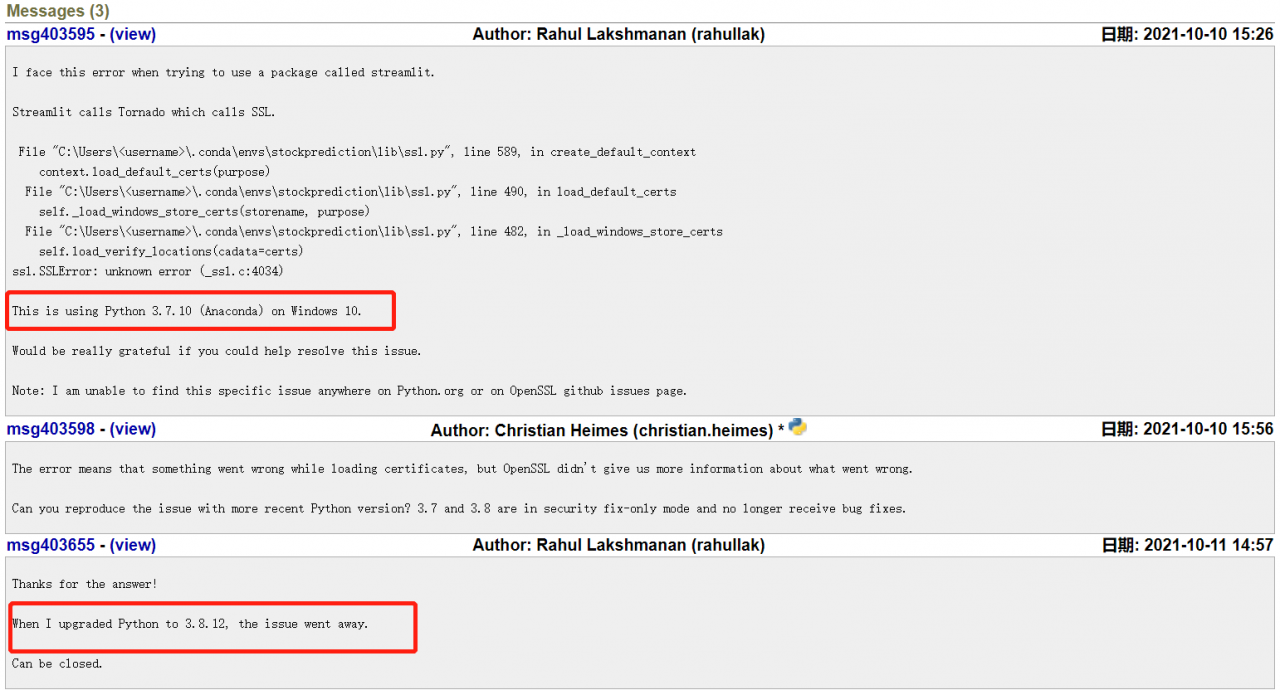 explanation on the Internet. I think it should be the python version, so I upgraded the python version in Anaconda navigator. Then enter
explanation on the Internet. I think it should be the python version, so I upgraded the python version in Anaconda navigator. Then enter 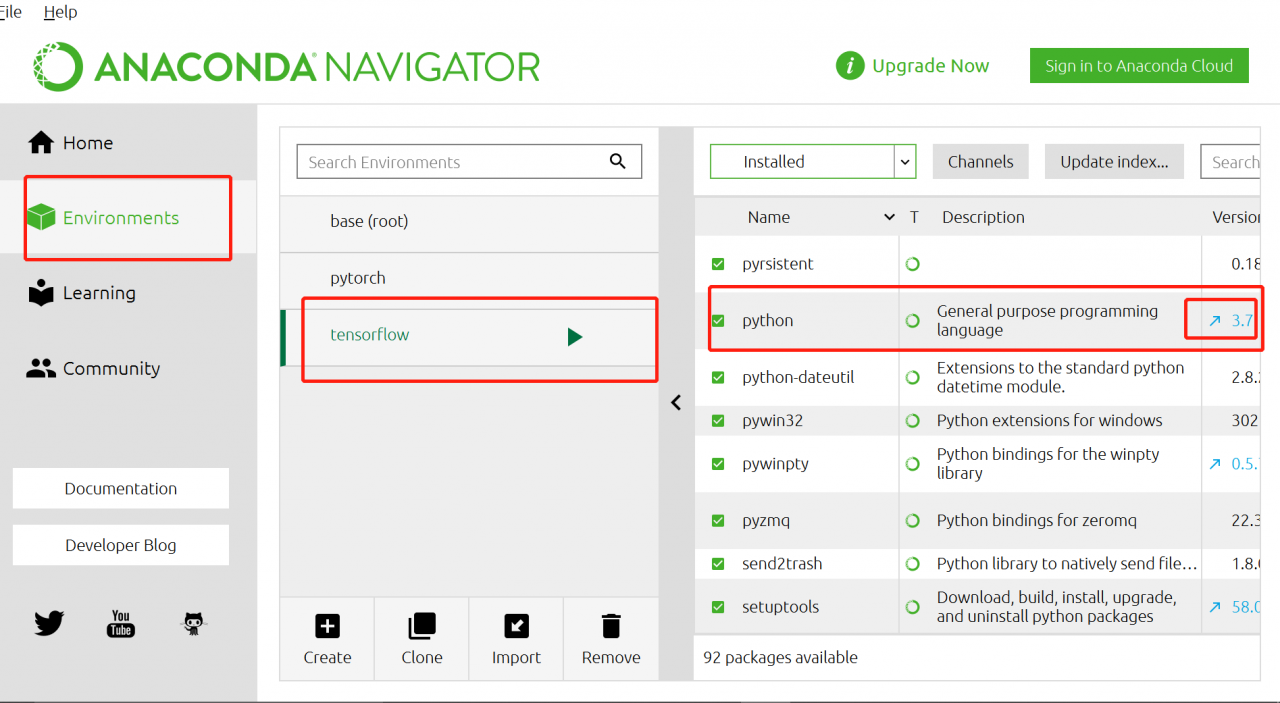
 .
.