1. Problem, the following problem occurred inexplicably. After tossing for a long time, I didn’t find out the reason. Later, I simply solved it by violence. I directly decompressed the file to see what happened. After decompressing for a long time, it prompted that the file was damaged,….
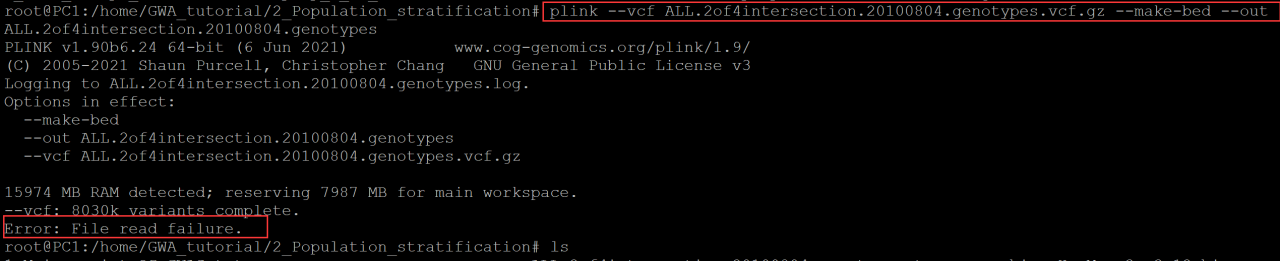
Error: File read failure.
2. Download the file again and test
root@PC1:/home/GWA_tutorial/2_Population_stratification#plink --vcf ALL.2of4intersection.20100804.genotypes.vcf.gz --make-bed --out ALL.2of4intersection.20100804.genotypes PLINK v1.90b6.24 64-bit (6 Jun 2021) www.cog-genomics.org/plink/1.9/ (C) 2005-2021 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to ALL.2of4intersection.20100804.genotypes.log. Options in effect: --make-bed --out ALL.2of4intersection.20100804.genotypes --vcf ALL.2of4intersection.20100804.genotypes.vcf.gz 15974 MB RAM detected; reserving 7987 MB for main workspace. --vcf: ALL.2of4intersection.20100804.genotypes-temporary.bed + ALL.2of4intersection.20100804.genotypes-temporary.bim + ALL.2of4intersection.20100804.genotypes-temporary.fam written. 25488488 variants loaded from .bim file. 629 people (0 males, 0 females, 629 ambiguous) loaded from .fam. Ambiguous sex IDs written to ALL.2of4intersection.20100804.genotypes.nosex . Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 629 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate is 0.615305. 25488488 variants and 629 people pass filters and QC. Note: No phenotypes present. --make-bed to ALL.2of4intersection.20100804.genotypes.bed + ALL.2of4intersection.20100804.genotypes.bim + ALL.2of4intersection.20100804.genotypes.fam ... done.
There was no error this time.
Error: file read failure error occurs when Plink processes large files One possibility is file corruption!