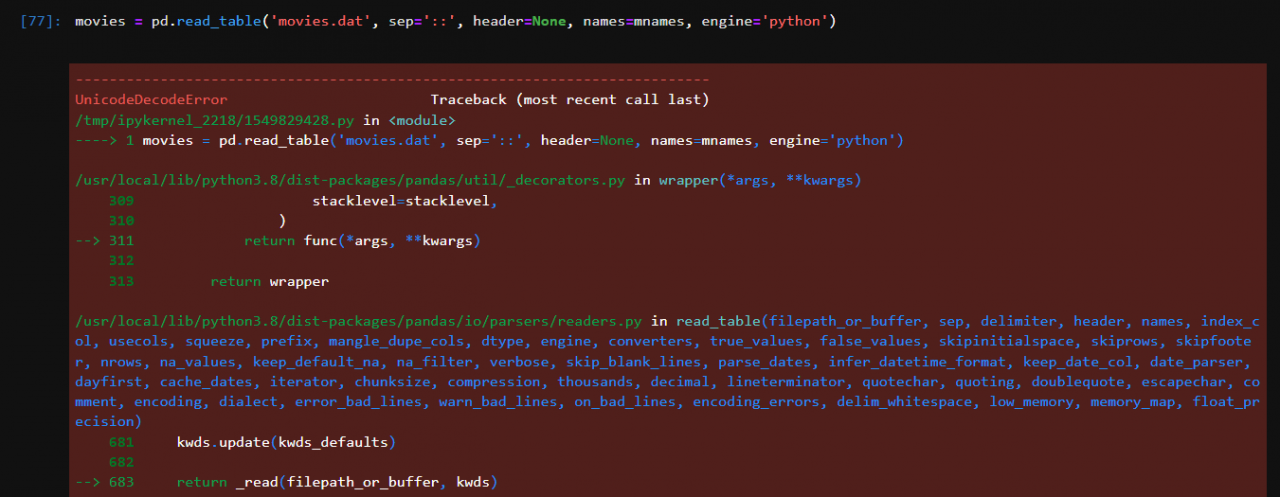
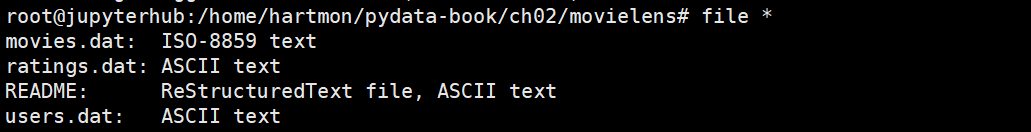
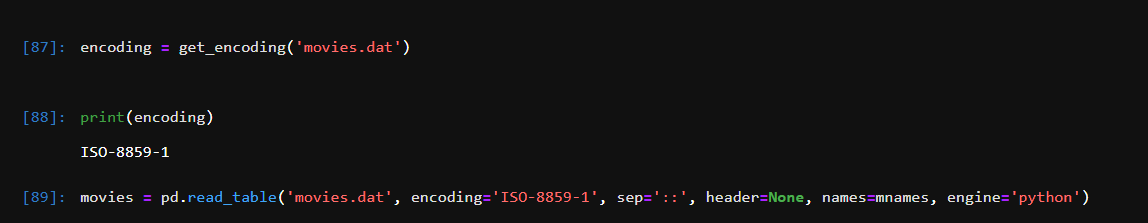
read_ excel()
The load function is read_ The specific parameters of Excel () are as follows
read_excel(io, sheetname=0, header=0, skiprows=None, skip_footer=0, index_col=None,names=None, parse_cols=None, parse_dates=False,date_parser=None,na_values=None,thousands=None, convert_float=True, has_index_names=None, converters=None,dtype=None, true_values=None, false_values=None, engine=None, squeeze=False, **kwds)
Common parameter analysis:
io : string, path object ; Excel path
sheetname: string, int, mixed list of Strings/ints, or none, default 0 returns multiple tables, use sheetname = [0,1], if sheetname = none returns the whole table, note: int/String returns dataframe, while none and list return Dict Of dataframe
header: int, list of ints, default 0 specifies the column name row, default 0, that is, take the first row, the data is the data below the column name row, if the data does not contain the column name, set header = none
skirows: list like, rows to skip at the beginning
skip_ Footer: int, default 0, omit the int line data from the tail
index_ Col: int, list of ints, default none specifies that the column is an index column. You can also use U “strings”
Names: array like, default none, specifies the name of the column
Data source:
sheet1:
ID NUM-1 NUM-2 NUM-3
36901 142 168 661
36902 78 521 602
36903 144 600 521
36904 95 457 468
36905 69 596 695
sheet2:
ID NUM-1 NUM-2 NUM-3
36906 190 527 691
36907 101 403 470
(1) Function prototype
basestation ="F://pythonBook_PyPDAM/data/test.xls"
data = pd.read_excel(basestation)
print data
Output: a dataframe
ID NUM-1 NUM-2 NUM-3
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695
(2) Sheetname parameter: use sheetname = [0,1] to return multiple tables. If sheetname = none, return the whole table. Note: int/String returns dataframe, while none and list return Dict Of dataframe
data_1 = pd.read_excel(basestation,sheetname=[0,1])
print data_1
print type(data_1)
Output: dict of dataframe
OrderedDict([(0, ID NUM-1 NUM-2 NUM-3
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695),
(1, ID NUM-1 NUM-2 NUM-3
0 36906 190 527 691
1 36907 101 403 470)])
(3) Header parameter: specify the row of column name, 0 by default, that is, the first row. The data is the data below the row of column name. If the data does not contain column name, set header = none. Note that there is a row of column name here
data = pd.read_excel(basestation,header=None)
print data
Output:
0 1 2 3
0 ID NUM-1 NUM-2 NUM-3
1 36901 142 168 661
2 36902 78 521 602
3 36903 144 600 521
4 36904 95 457 468
5 36905 69 596 695
data = pd.read_excel(basestation,header=[3])
print data
Output:
36903 144 600 521
0 36904 95 457 468
1 36905 69 596 695
(4) Skirows parameter: omits the data with the specified number of rows
data = pd.read_excel(basestation,skiprows = [1])
print data
Output:
ID NUM-1 NUM-2 NUM-3
0 36902 78 521 602
1 36903 144 600 521
2 36904 95 457 468
3 36905 69 596 695
(5)skip_ Footer parameter: omits the data from the int line of the tail number
data = pd.read_excel(basestation, skip_footer=3)
print data
Output:
ID NUM-1 NUM-2 NUM-3
0 36901 142 168 661
1 36902 78 521 602
(6)index_ Col parameter: specify column as index column, or use U “strings”
data = pd.read_excel(basestation, index_col="NUM-3")
print data
output:
ID NUM-1 NUM-2
NUM-3
661 36901 142 168
602 36902 78 521
521 36903 144 600
468 36904 95 457
695 36905 69 596
(7) Name parameter: Specifies the name of the column
data = pd.read_excel(basestation,names=["a","b","c","e"])
print data
a b c e
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695
The specific parameters are as follows
>>> print help(pandas.read_excel)
Help on function read_excel in module pandas.io.excel:
read_excel(io, sheetname=0, header=0, skiprows=None, skip_footer=0, index_col=None, names=None, parse_cols=None, parse_dates=False, date_parser=None, na_values=None, thousands=None, convert_float=True, has_index_names=None, converters=None, dtype=None, true_values=None, false_values=None, engine=None, squeeze=False, **kwds)
Read an Excel table into a pandas DataFrame
Parameters
----------
io : string, path object (pathlib.Path or py._path.local.LocalPath),
file-like object, pandas ExcelFile, or xlrd workbook.
The string could be a URL. Valid URL schemes include http, ftp, s3,
and file. For file URLs, a host is expected. For instance, a local
file could be file://localhost/path/to/workbook.xlsx
sheetname : string, int, mixed list of strings/ints, or None, default 0
Strings are used for sheet names, Integers are used in zero-indexed
sheet positions.
Lists of strings/integers are used to request multiple sheets.
Specify None to get all sheets.
str|int -> DataFrame is returned.
list|None -> Dict of DataFrames is returned, with keys representing
sheets.
Available Cases
* Defaults to 0 -> 1st sheet as a DataFrame
* 1 -> 2nd sheet as a DataFrame
* "Sheet1" -> 1st sheet as a DataFrame
* [0,1,"Sheet5"] -> 1st, 2nd & 5th sheet as a dictionary of DataFrames
* None -> All sheets as a dictionary of DataFrames
header : int, list of ints, default 0
Row (0-indexed) to use for the column labels of the parsed
DataFrame. If a list of integers is passed those row positions will
be combined into a ``MultiIndex``
skiprows : list-like
Rows to skip at the beginning (0-indexed)
skip_footer : int, default 0
Rows at the end to skip (0-indexed)
index_col : int, list of ints, default None
Column (0-indexed) to use as the row labels of the DataFrame.
Pass None if there is no such column. If a list is passed,
those columns will be combined into a ``MultiIndex``. If a
subset of data is selected with ``parse_cols``, index_col
is based on the subset.
names : array-like, default None
List of column names to use. If file contains no header row,
then you should explicitly pass header=None
converters : dict, default None
Dict of functions for converting values in certain columns. Keys can
either be integers or column labels, values are functions that take one
input argument, the Excel cell content, and return the transformed
content.
dtype : Type name or dict of column -> type, default None
Data type for data or columns. E.g. {'a': np.float64, 'b': np.int32}
Use `object` to preserve data as stored in Excel and not interpret dtype.
If converters are specified, they will be applied INSTEAD
of dtype conversion.
.. versionadded:: 0.20.0
true_values : list, default None
Values to consider as True
.. versionadded:: 0.19.0
false_values : list, default None
Values to consider as False
.. versionadded:: 0.19.0
parse_cols : int or list, default None
* If None then parse all columns,
* If int then indicates last column to be parsed
* If list of ints then indicates list of column numbers to be parsed
* If string then indicates comma separated list of Excel column letters and
column ranges (e.g. "A:E" or "A,C,E:F"). Ranges are inclusive of
both sides.
squeeze : boolean, default False
If the parsed data only contains one column then return a Series
na_values : scalar, str, list-like, or dict, default None
Additional strings to recognize as NA/NaN. If dict passed, specific
per-column NA values. By default the following values are interpreted
as NaN: '', '#N/A', '#N/A N/A', '#NA', '-1.#IND', '-1.#QNAN', '-NaN', '-nan',
'1.#IND', '1.#QNAN', 'N/A', 'NA', 'NULL', 'NaN', 'nan'.
thousands : str, default None
Thousands separator for parsing string columns to numeric. Note that
this parameter is only necessary for columns stored as TEXT in Excel,
any numeric columns will automatically be parsed, regardless of display
format.
keep_default_na : bool, default True
If na_values are specified and keep_default_na is False the default NaN
values are overridden, otherwise they're appended to.
verbose : boolean, default False
Indicate number of NA values placed in non-numeric columns
engine: string, default None
If io is not a buffer or path, this must be set to identify io.
Acceptable values are None or xlrd
convert_float : boolean, default True
convert integral floats to int (i.e., 1.0 --> 1). If False, all numeric
data will be read in as floats: Excel stores all numbers as floats
internally
has_index_names : boolean, default None
DEPRECATED: for version 0.17+ index names will be automatically
inferred based on index_col. To read Excel output from 0.16.2 and
prior that had saved index names, use True.
Returns
to_ excel()
The storage function is pd.dataframe.to_ Excel (), note that dataframe must be written into excel, that is, write dataframe to an excel sheet. The specific parameters are as follows:
to_excel(self, excel_writer, sheet_name='Sheet1', na_rep='', float_format=None,columns=None, header=True, index=True, index_label=None,startrow=0, startcol=0, engine=None, merge_cells=True, encoding=None,
inf_rep='inf', verbose=True, freeze_panes=None)
Explanation of common parameters:
excel_writer: string or ExcelWriter object File path or existing ExcelWriter Target Path
sheet_name: string, default ‘Sheet1’ Name of sheet which will contain DataFrame, Fill the pages of excel
na_rep: string, default ”,Missing data representation Missing value fill
float_format: string, default None Format string for floating point numbers
columns: sequence, optional,Columns to write Select the output column.
header: boolean or list of string, default True Write out column names. If a list of string is given it is assumed to be aliases for the column names
index: boolean, default True,Write row names (index)
index_label : string or sequence, default None, Column label for index column(s) if desired. If None is given, andheader and index are True, then the index names are used. A sequence should be given if the DataFrame uses MultiIndex.
startrow: upper left cell row to dump data frame
startcol: upper left cell column to dump data frame
engine: string, default None ,write engine to use – you can also set this via the options,io.excel.xlsx.writer, io.excel.xls.writer, andio.excel.xlsm.writer.
merge_cells: boolean, default True Write MultiIndex and Hierarchical Rows as merged cells.
encoding: string, default None encoding of the resulting excel file. Only necessary for xlwt,other writers support unicode natively.
inf_rep: string, default ‘inf’ Representation for infinity (there is no native representation for infinity in Excel)
freeze_panes: tuple of integer (length 2), default None Specifies the one-based bottommost row and rightmost column that is to be frozen
Codes:
ID NUM-1 NUM-2 NUM-3
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695
5 36906 165 453
datas:
basestation ="F://python/data/test.xls"
basestation_end ="F://python/data/test_end.xls"
data = pd.read_excel(basestation)
(1) Parameter Excel_ Writer, output path
data.to_excel(basestation_end)
Output:
ID NUM-1 NUM-2 NUM-3
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695
5 36906 165 453
(2)sheet_ Name, store the data in the sheet page of Excel
data.to_excel(basestation_end,sheet_name="sheet2")
(3)na_ Rep, missing value filling
data.to_excel(basestation_end,na_rep="NULL")
Output:
ID NUM-1 NUM-2 NUM-3
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695
5 36906 165 453 NULL
(4) Columns parameters: sequence, optional, columns to write select the output column
data.to_excel(basestation_end,columns=["ID"])
Output:
ID
0 36901
1 36902
2 36903
3 36904
4 36905
5 36906
(5) Header parameter: Boolean or list of string, which is true by default. You can use list to name columns. If header = false, no header will be output
data.to_excel(basestation_end,header=["a","b","c","d"])
Output:
a b c d
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695
5 36906 165 453
data.to_excel(basestation_end,header=False,columns=["ID"])
header = False
Output:
0 36901
1 36902
2 36903
3 36904
4 36905
5 36906
(6) Index: Boolean, default true, write row names (index) is true by default, and the index is displayed. When index = false, the row index (name) is not displayed. index_ Label: string or sequence, default none set the column name of the index column
data.to_excel(basestation_end,index=False)
Output:
ID NUM-1 NUM-2 NUM-3
36901 142 168 661
36902 78 521 602
36903 144 600 521
36904 95 457 468
36905 69 596 695
36906 165 453
data.to_excel(basestation_end,index_label=["f"])
Output:
f ID NUM-1 NUM-2 NUM-3
0 36901 142 168 661
1 36902 78 521 602
2 36903 144 600 521
3 36904 95 457 468
4 36905 69 596 695
5 36906 165 453